The Yeast Genomics* lab @ NOVA
(*evolutionary, functional, comparative and more)




The beginning of agriculture and the domestication of plants and animals are among the most decisive events in the past 13,000 years of human history. Archaeological and biomolecular evidence indicates that rice wine was produced as far back as 7,000 BC in China. In Zagros mountains in Iran, wine was produced 5,000 BC and the domestication of barley in the Fertile Crescent led to the emergence of the forebear of modern beer in Sumeria 4,000 BC. Furthermore, yeast cells were detected in Egyptian leavened bread dough dated 3,000 BC and molecular evidence for the presence of Saccharomyces cerevisiae in wine fermentation has been obtained from pottery jars of the same period.
Ethanol and many other metabolites contributed to preserve dietary goods and opened the way for the production of a myriad of alcoholic beverages that become important in social habits of many civilizations.
In spite of their long association to the fermentation of foods and beverages, the natural biology of Saccharomyces remains elusive. Little is known about the ecology of the seven natural species and their biology outside the laboratory still has many mysteries.
Through extensive field studies in temperate oak forests in different continents and equivalent biotopes in the Southern Hemisphere we aim at a better understanding of the natural history of Saccharomyces. We are specifically interested in discovering wild populations and to perform whole-
Yeast Evolutionary Ecology and Domestication
José Paulo Sampaio
We reported in PNAS the identification in Patagonia of Saccharomyces eubayanus, one of the ancestors of the complex hybrid genome of lager yeast (Saccharomyces pastorianus). This study involved the isolation of dozens of strains of S. eubayanus and its sister species S. uvarum from Nothofagus (Southern beech) and Cyttaria, its biotrophic fungal parasite. We detected S. eubayanus and S. uvarum in sympatry, but found that the two species were genetically isolated through a post-
(portuguese press coverage of this research here)
Detection of Saccharomyces eubayanus in Patagonia

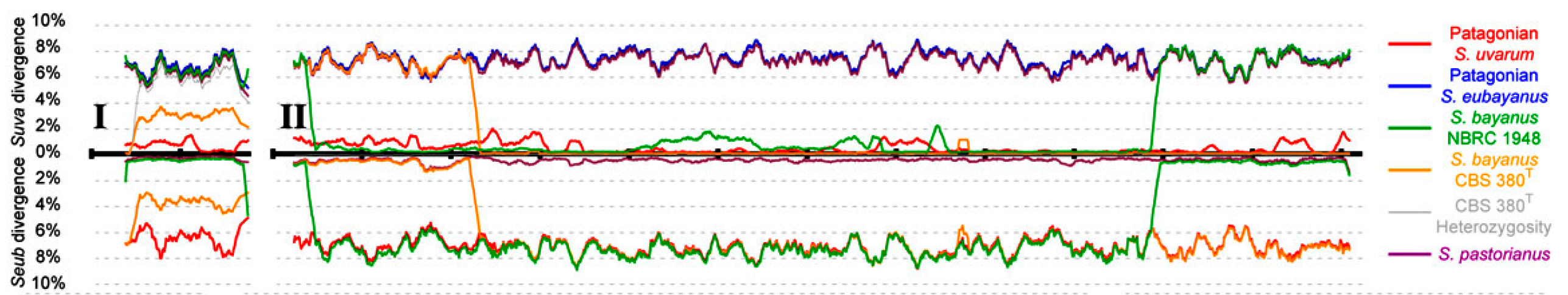
S. eubayanus sp. nov. is distinct from S. uvarum and donated its genome to S. pastorianus.
A model of the formation of S. pastorianus and the hybrid strains of S. bayanus
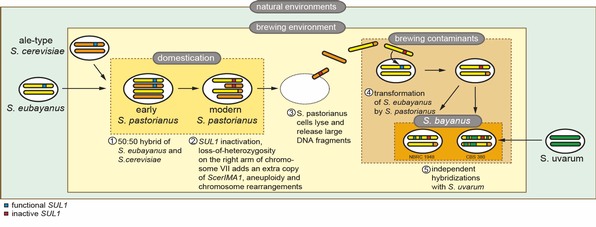


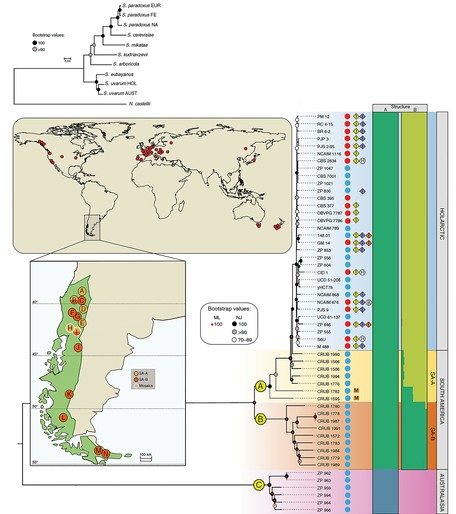
We reported in Nature Communications a population genomics study of the cider and wine yeast S. uvarum using the most comprehensive strain dataset currently available and involving new isolates collected by us in New Zealand, Tasmania, and in South and North America. We revealed a highly diverse South American population containing more genetic diversity than that found in the Northern Hemisphere. Moreover we documented that a Patagonian sub-
The global phylogeography and the first indication of domestication in Saccharomyces uvarum


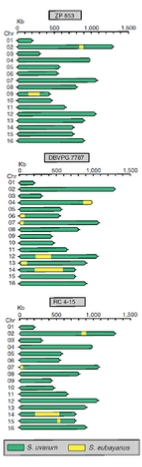
Geographic distribution, phylogeny and population structure of S. uvarum
Chromosome maps of S. uvarum strains showing the location and extent of introgressions from S. eubayanus



The Mediterranean origins of wine yeast domestication
The domestication of the wine yeast Saccharomyces cerevisiae is thought to be contemporary with the development of viticulture along the Mediterranean basin. Until recently, the unavailability of wild lineages prevented the identification of the closest wild relatives of wine yeasts. To bridge this gap, we used whole-
(read the accompanied commentary here)
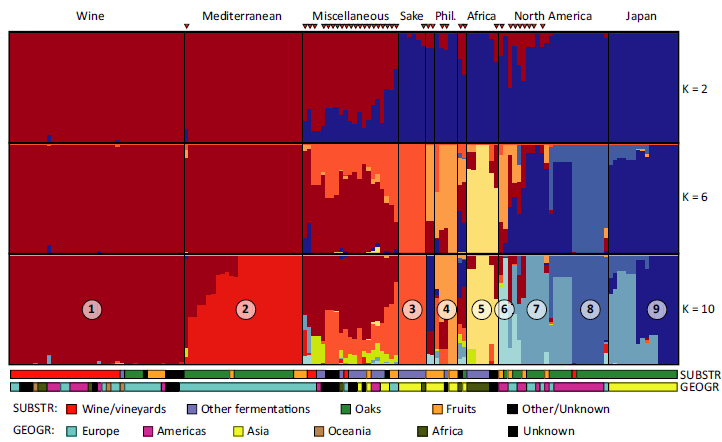
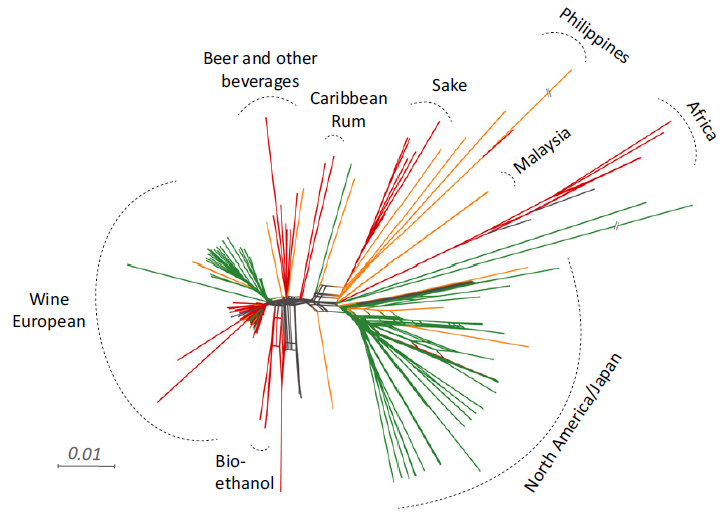
Population structure and admixture in Saccharomyces cerevisiae
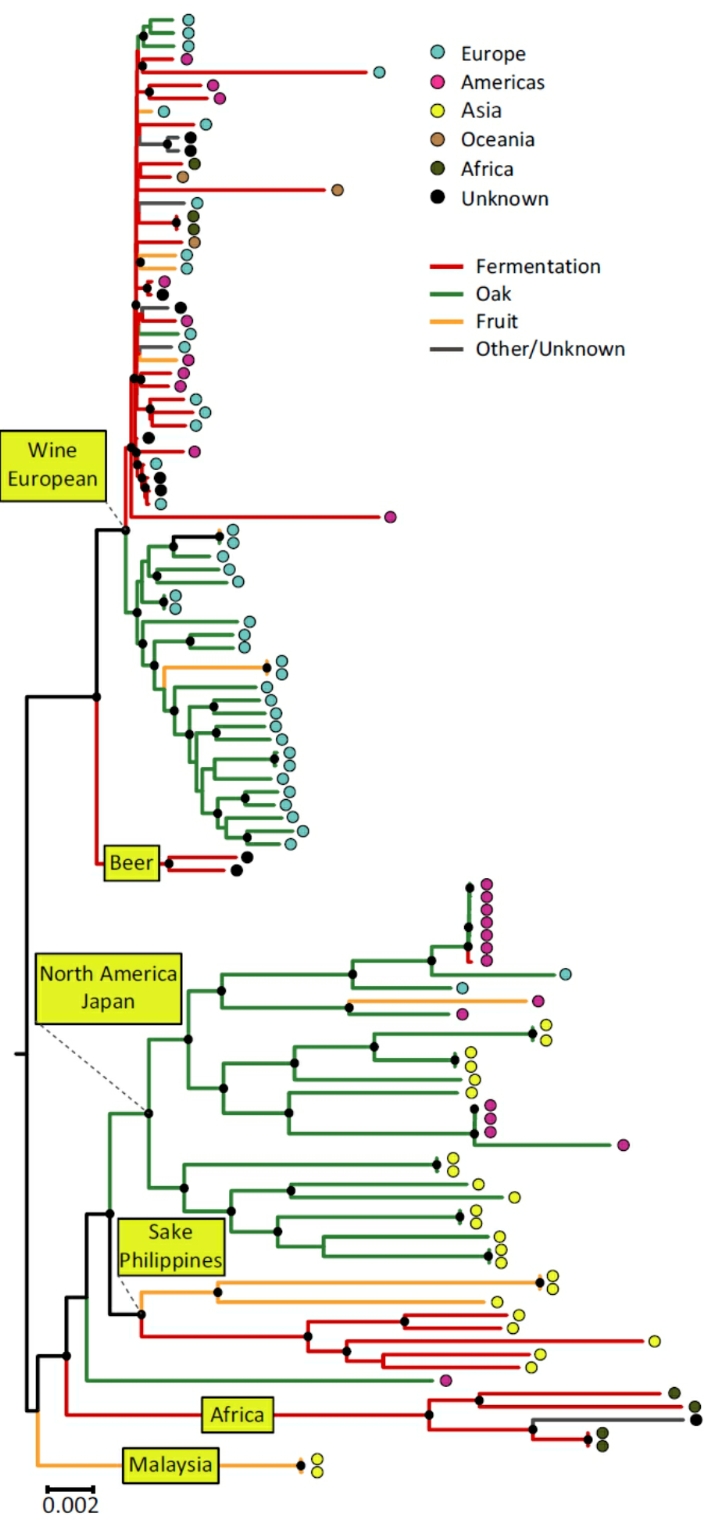
The global diversity of Saccharomyces cerevisiae is shaped by both ecology and geography. Neighbour-
Whole-

Natural hybridization in Brazilian wild lineages of S. cerevisiae
The global distribution of natural populations of S. cerevisiae, especially in tropical regions such as Brazil were oaks and other members of the Fagaceae are absent, is not well understood. In the work recently published in Genome Biology and Evolution we investigated the occurrence of S. cerevisiae in Brazil, and reported a candidate natural habitat of S. cerevisiae in South America. Using whole-
We hypothesize that hybridization in tropical wild lineages may have facilitated the habitat transition accompanying the colonization of the tropical ecosystem.
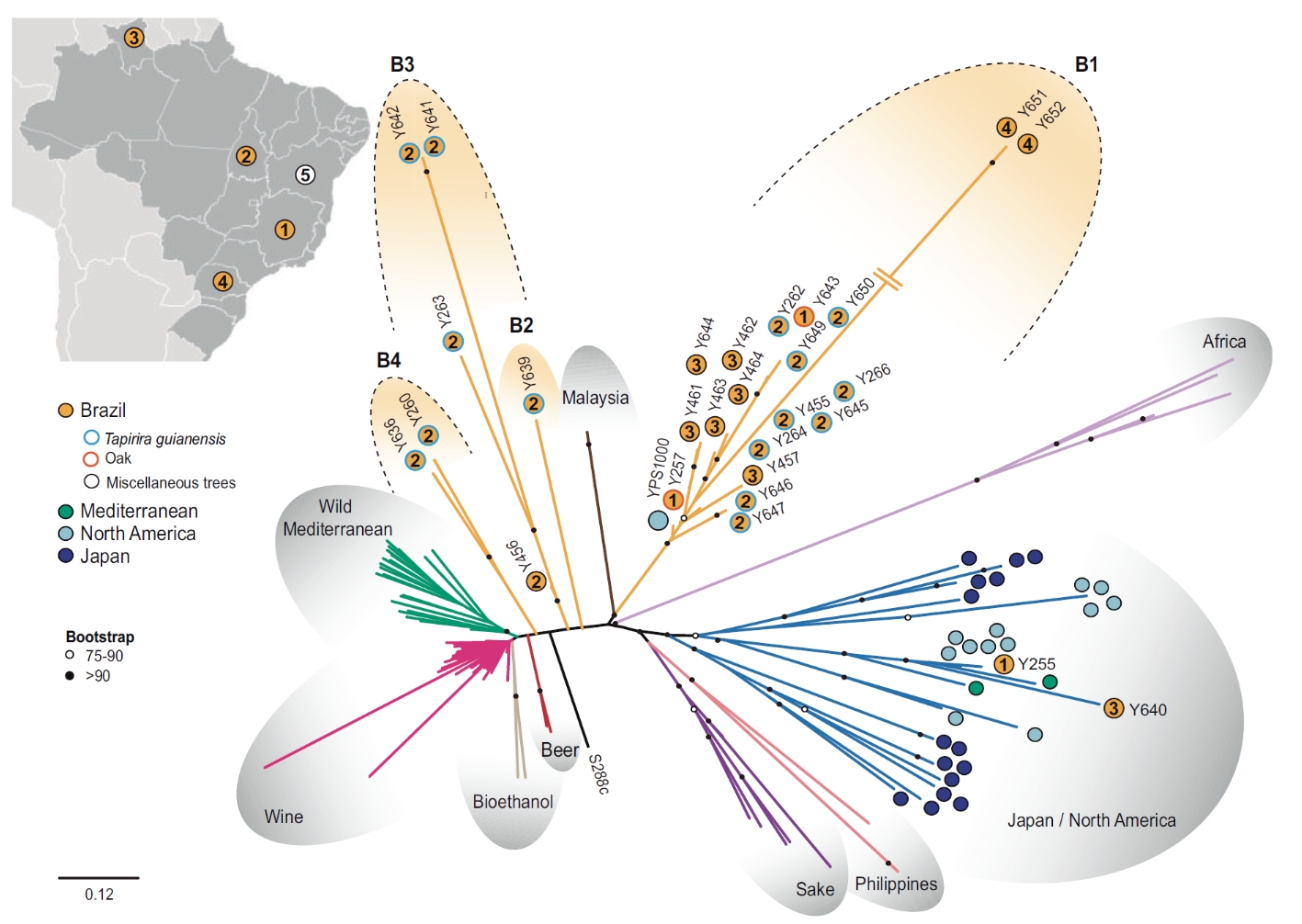
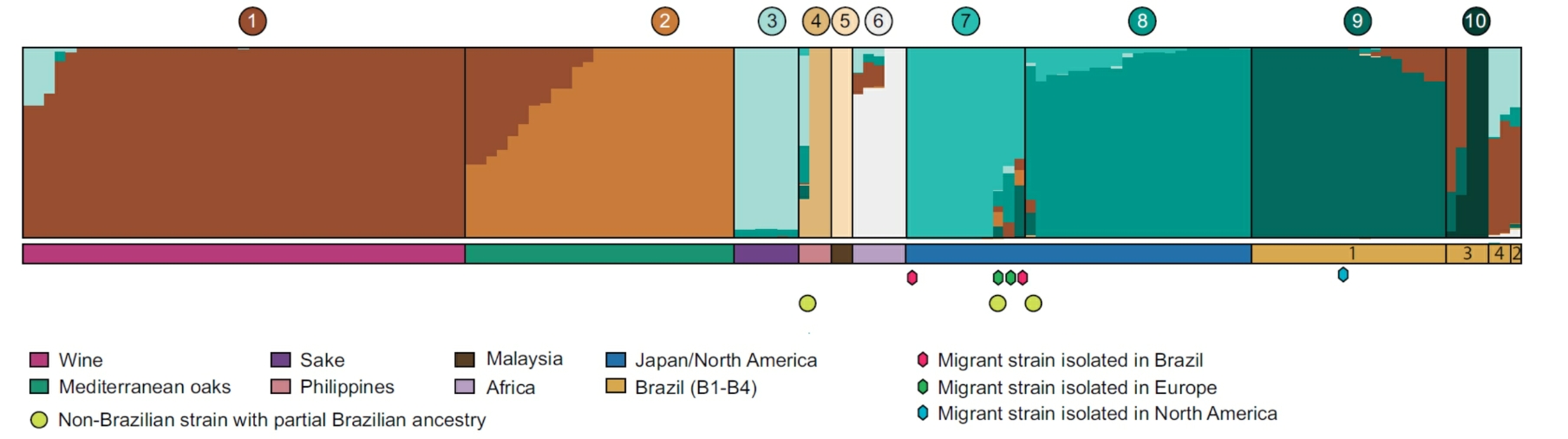
Population structure of S. cerevisiae. Migrant strains are marked with colored diamonds: red (isolated in Brazil), green (isolated in Europe), and blue (isolated in North America). Yellow circles depict non-
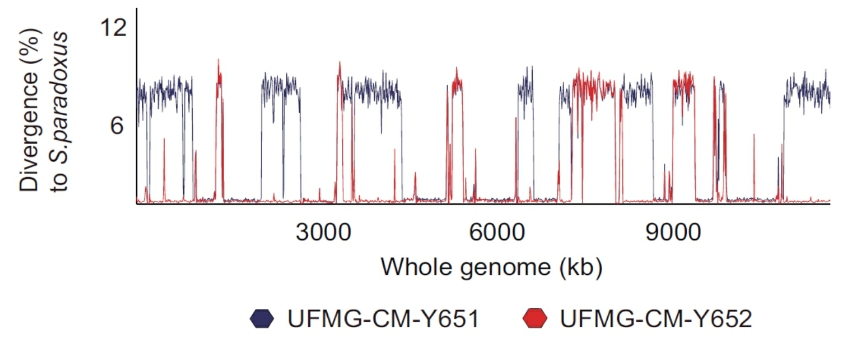
Saccharomyces cerevisiae X S. paradoxus hybrid genomes
Whole-