|
Phenotypic
evolution and ecological speciation in
Saccharomyces sensu stricto
Speciation is one of the least understood major features of evolution
because of the number and complexity of the mechanisms that might lead to the
evolution of distinct species. We are using the species of the genus Saccharomyces
to understand how evolution shaped their (very similar) phenotypes and to
understand why various Saccharomyces
species are able to co-exist in the same habitat. We are combining genome,
ecological and physiological data to test our hypotheses on the major
evolutionary trends that drove speciation within this genus.

|
|
|
The yeasts belonging to the genus Saccharomyces,
especially S. cerevisiae, play an important role in human activities.
They are used as fermenting agents worldwide and stand out as eukaryotic
model organisms.
|
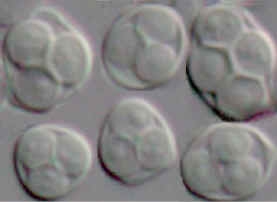
Asci and ascospores of Saccharomyces
kudriavzevii ZP 591.
© J.P. Sampaio 2008
|
|
|
In spite of the extensive research on Saccharomyces
yeasts, little is known about their natural habitats and population
genetics. Traditionally S. paradoxus has been regarded as a wild
species associated mostly with natural habitats. On the contrary, several
authors viewed S. cerevisiae as a domesticated organism chiefly
adapted to man-made fermentations and normally absent in natural ecosystems.
However, several lines of evidence suggest that S. cerevisiae existed
in natural environments long before it was utilized in man-made
fermentations. Because ecological data on Saccharomyces is fragmentary and in some cases is based on erroneous
species identifications, the habitats of most species remain unknown.
Recently, we found an unprecedented diversity of Saccharomyces sensu
stricto species associated with oak trees in Portugal. Among these, a
European population of S. kudriavzevii, a species thus far thought to
be endemic of Japan, was identified.
|

Sampling site at Castelo de Vide, Portugal.
© J.P. Sampaio 2008
|
|

Isolation of Saccharomyces
spp. from oak bark – enrichment step.
© J.P. Sampaio 2008
|
|
|
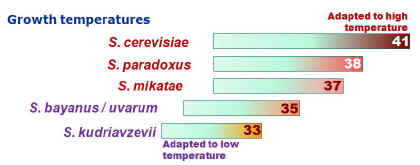
Interestingly, at all the localities where S. kudriavzevii was
found, it co-existed with either S. cerevisiae or S. paradoxus.
Saccharomyces kudriavzevii, like S. uvarum (formerly known as S.
bayanus var. uvarum), exhibited a strong preference for lower
growth temperatures when compared with S. cerevisiae and S.
paradoxus.
|
|
|

|
|
|
Temperature adaptation and ecological
speciation in Saccharomyces
Our general goal is to understand how the
various Saccharomyces species
evolved from their primal Saccharomyces
ancestor and what forces drove speciation in this group. We are also
interested in understanding the particularities of each present-day species
i.e., we would like to know the set of species-specific characteristics -
those phenotypic traits that make each species unique. This issue is of
particular relevance because (i) the phenotypes of the different Saccharomyces
species are very similar, and (ii) according to the principle of niche
exclusion, if more than one species exist in the same ecological niche,
competition will occur and one species will outcompete and exclude the
others. Therefore, if more than one Saccharomyces
species are found in the same habitat, what kinds of ecological speciation
evens have taken place during the course of evolution?
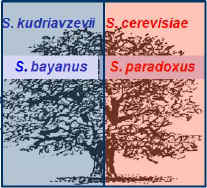
We hypothesize that temperature adaptation played a crucial role in the
phenotypic evolution of sympatric Saccharomyces sensu stricto
species. This would explain why S. cerevisiae (thermophilic) and S.
kudriavzevii (cryotolerant) are able to apparently evade the competitive
exclusion principle and to consistently establish sympatric associations on
tree bark.
|
|
|

|
|
|
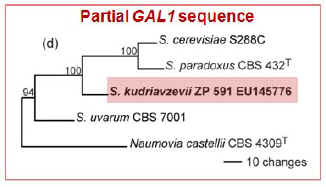
Galactose utilization in S.
kudriavzevii
Prior to this study, only four S.
kudriavzevii strains were known and
all exhibited one important trait that distinguished them from other Saccharomyces
species: they were unable to grow on galactose. We observed that the
Portuguese S. kudriavzevii population exhibits important genetic differences
when compared with the strains of this species previously known. The
most striking difference is undoubtedly the presence of functional GAL
genes in the Portuguese population. It was shown that the inability of the
type strain of S. kudriavzevii to grow on galactose was due to
extensive and ancient gradual degeneration of the entire set of genes
involved in galactose utilization, rather than from a discrete mutation
event limited in time that could have taken place very recently (Hittinger
et al. 2004). Calculations taking
into account the neutral mutation rates in S.
kudriavzevii, date GAL pathway degeneration to a period immediately
following separation of the lineage leading to S.
kudriavzevii. This is difficult to reconcile with the fact that the
Portuguese isolates have otherwise only modest sequence divergence with
respect to the type strain.
|
|
|

|
|
|
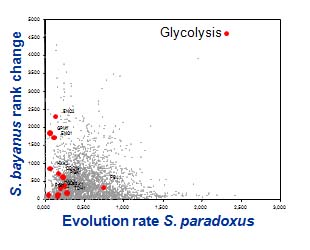
Genomes and the evolution of phenotypes
We are using a genomic approach aiming at identifying genes likely to be
involved in phenotypic evolution via disruptive selection and character
displacement. The ratios of nonsynonymous versus synonymous nucleotide
substitutions were used to perform genome-level pairwise comparisons of
genetic differences. We found that while most genes appear to evolve at a
similar pace in all species, some exhibit striking differences between the
different genomes studied. This analysis allowed us to highlight a set of
proteins that is significantly more divergent in S. kudriavzevii and/or
S. uvarum than in S. paradoxus, when these genomes are compared
to that of S. cerevisiae. Within this set of divergent proteins, some
functional classes were found to be particularly well represented.
|
|
|
|
|
|
|